AVIAN INFLUENZA:
DO WE NEED TO WORRY ABOUT THE LATEST BIRD FLU OUTBREAKS?
By: Yan Mardian
An 11-year-old girl has died of bird flu in Cambodia, south-east Asia, and multiple others who live in her area have been sickened, including her father, according to media reports, marking the first known H5N1 human infections in the country since 2014—and potentially setting the stage for sustained human-to-human transmission. The cases raised fears that the virus had acquired the ability to spread among people and may trigger another pandemic. But the World Health Organization said that 11 contacts of the girl, four of whom have flulike symptoms, had tested negative for infection with the H5N1 flu virus. Cambodian authorities said that the deceased girl and survived father was infected with an older viral variant H5 clade 2.3.2.1c which has circulated in Cambodia among birds/poultry for many years and has sporadically caused human infection. The virus is unrelated to the outbreaks in birds caused by a new strain of H5N1, clade 2.3.4.4b, which emerged in 2020, in the United States and Europe. While these avian Influenza viruses can cause human infections, it has not been seen to cause human-to-human transmission to date.
Influenza virus
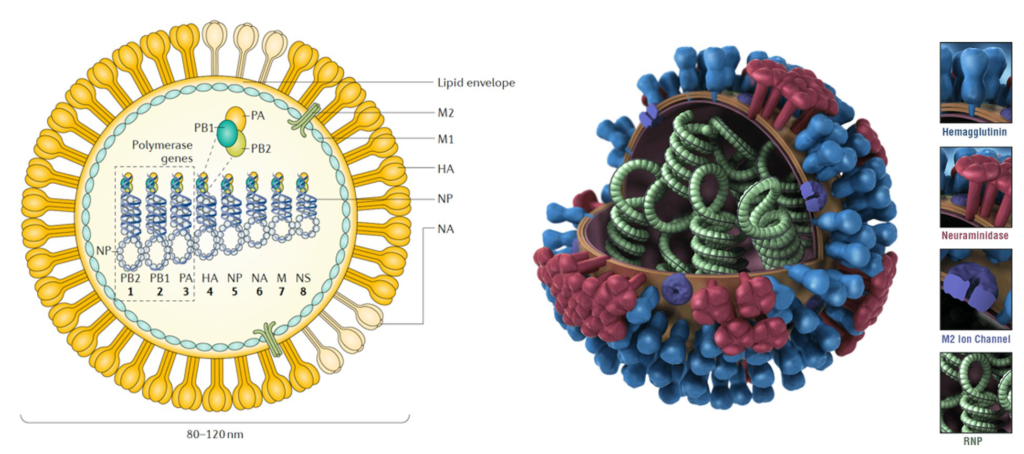
Influenza is an infectious respiratory disease; in humans, it is caused by influenza A (genus influenza virus A) and influenza B (genus influenza virus B) viruses (influenza virus C and influenza virus D genera are also known). All influenza viruses are enveloped negative-sense single-strand RNA viruses with a segmented genome. Influenza A and B viruses contain eight RNA segments, which encode RNA polymerase subunits, viral glycoproteins (namely, haemagglutinin (HA), with its distinct globular ‘head’ and ‘stalk’ structures, which facilitate viral entry, and neuraminidase (NA), which facilitates viral release), viral nucleoprotein (NP), matrix protein (M1) and membrane protein (M2), the nonstructural protein NS1 and nuclear export protein (NEP). The HA and NA viral proteins are the most antigenically variable, and in the case of influenza A virus, they are classified into antigenically diverse subtypes. These two viral glycoproteins are located at the surface of the virus particle and are the main targets for protective antibodies induced by influenza virus infection and vaccination.
The segmented nature of the influenza viral genome enables reassortment, that is, interchange, of genomic RNA segments when two viruses of the same type (that is, two influenza A viruses or two influenza B viruses) infect the same cell. A unique characteristic of influenza A viruses is that they circulate not only in humans but also in domestic animals, pigs, horses and poultry and in wild migratory birds (>100 species of ducks, geese, swans, gulls, waders and wild aquatic birds are considered natural reservoirs. These animal reservoirs provide a source of antigenically diverse HA and NA genes that can be exchanged between viral strains by reassortment after co-infection of the same host, increasing virus diversity and in some instances leading to the generation of human pandemic influenza virus strains with HA and/or NA derived from animal strains. By contrast, influenza B and influenza C viruses are not divided into different subtypes and are restricted to humans, with no known animal reservoirs, although limited spillover to seals and pigs has occurred, respectively.
Avian influenza virus
Avian influenza (AI) viruses are highly contagious, extremely variable viruses that are widespread in birds. Wild birds in aquatic habitats are thought to be their natural reservoir hosts, but domesticated poultry and other birds can also be infected. Most viruses cause only mild disease in poultry and are called low pathogenic avian influenza (LPAI) viruses. Avian influenza viruses can occasionally affect mammals, including humans, usually after close contact with infected poultry. While infections in people are often limited to conjunctivitis or mild respiratory disease, some viruses can cause severe illness. During the last century, such viruses have caused or contributed to at least three pandemics in humans, contributed to the diversity of swine influenza viruses in pigs, and produced one of the two canine influenza viruses now circulating among dogs. AI viruses belonging to the species influenza A virus, genus influenza virus A and family Orthomyxoviridae. Influenza A viruses are classified into subtypes based on two surface proteins, the hemagglutinin (HA) and neuraminidase (NA). A virus that has a type 1 HA and type 2 NA, for example, would have the subtype H1N2. At least 16 hemagglutinins (H1 to H16), and 9 neuraminidases (N1 to N9) have been found in viruses from birds.
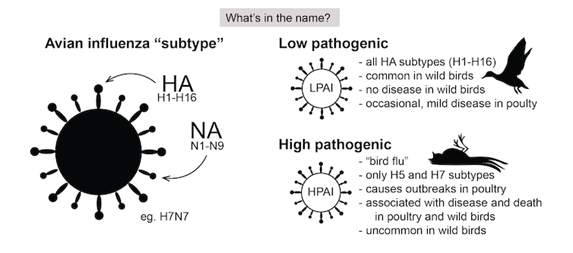
Avian influenza A viruses are classified into the following two categories: low pathogenicity avian influenza (LPAI) A viruses, and highly pathogenic avian influenza (HPAI) A viruses. The categories refer to molecular characteristics of a virus and the virus’ ability to cause disease and mortality in chickens in a laboratory setting. Highly pathogenic avian influenza (HPAI) viruses can develop from certain LPAI viruses, usually while they are circulating in poultry flocks. HPAI viruses can kill up to 90-100% of the flock, and cause epidemics that may spread rapidly. HPAI and LPAI are defined and explained below:
- Low Pathogenic Avian Influenza (LPAI): Low pathogenic avian influenza viruses cause either no signs of disease or mild disease in chickens/poultry. Most avian influenza A viruses are low pathogenic and cause few signs of disease in infected wild birds. In poultry, some LPAI viruses can mutate into HPAI avian influenza viruses.
- Highly Pathogenic Avian Influenza (HPAI): Highly pathogenic avian influenza viruses cause severe dis-ease and high mortality in infected poultry. Only some avian influenza A(H5) and A(H7) viruses are classified as HPAI A viruses, while most A(H5) and A(H7) viruses circulating among birds are LPAI A viruses. HPAI A(H5) or A(H7) virus infections can cause disease that affects multiple internal organs with mortality up to 90% to 100% in chickens, often within 48 hours. HPAI A(H5) and A(H7) virus infections in poultry also can spill back into wild birds, resulting in further geographic spread of the virus as those birds migrate. While some wild bird species can be infected with some HPAI A(H5) or A(H7) virus sub-types without appearing sick, other HPAI A(H5) and A(H7) subtypes can cause severe disease and mortality in some infected wild birds/poultry.
HPAI and LPAI designations do not refer to or correlate with the severity of illness in cases of human infection with these viruses; both LPAI and HPAI A viruses have caused mild to severe illness in infected humans. There are genetic and antigenic differences between the influenza A virus subtypes that typically infect only birds and those that can infect birds and people.
The first known human cases of AI were reported in China and Hong Kong in 1997, where transmission from animals to humans led to 18 people being infected, of whom six died. Since then 19 countries have reported more than 860 H5N1 human infections to the World Health Organization from 2003 to 2022. Of these, 53% have resulted in death. Since 2005, HPAI A(H5N1) viruses have undergone extensive genetic diversification including the formation of hundreds of genotypes following reassortment with other avian influenza A viruses. Clade 2.3.4.4b HPAI A(H5N1) viruses emerged in 2020 and were introduced into North America in late 2021 and have spread to Central and South America, resulting in wild bird and poultry outbreaks in many countries. Globally, this 2.3.4.4b clade of HPAI A(H5N1) viruses has become widespread causing record numbers of bird outbreaks. Recently, over 11,300 animal outbreaks of HPAI A(H5N1) viruses were reported by 73 member states to the World Organization for Animal Health since January 2022.
While HPAI A(H5N1) viruses are currently circulating widely in wild birds and poultry in many geographic regions, relatively few human cases of A(H5N1) have been reported in recent years. Between January 2022 and March 15, 2023, only ten sporadic human cases of A(H5N1) were reported from seven countries. All reported cases had recent exposure to sick or dead poultry, and no cases of human-to-human HPAI A(H5N1) virus trans-mission were identified. Five cases (3 children, 2 adults) had severe disease, and 2 died. Seven cases were associated with clade 2.3.4.4b HPAI A(H5N1) viruses, and two cases were associated with clade 2.3.2.1c HPAI A(H5N1) viruses (the ones mentioned earlier); none of the HPAI A(H5N1) virus genetic sequences contained any known markers of reduced susceptibility to currently recommended FDA-approved influenza antiviral medications.
Antigenic drift
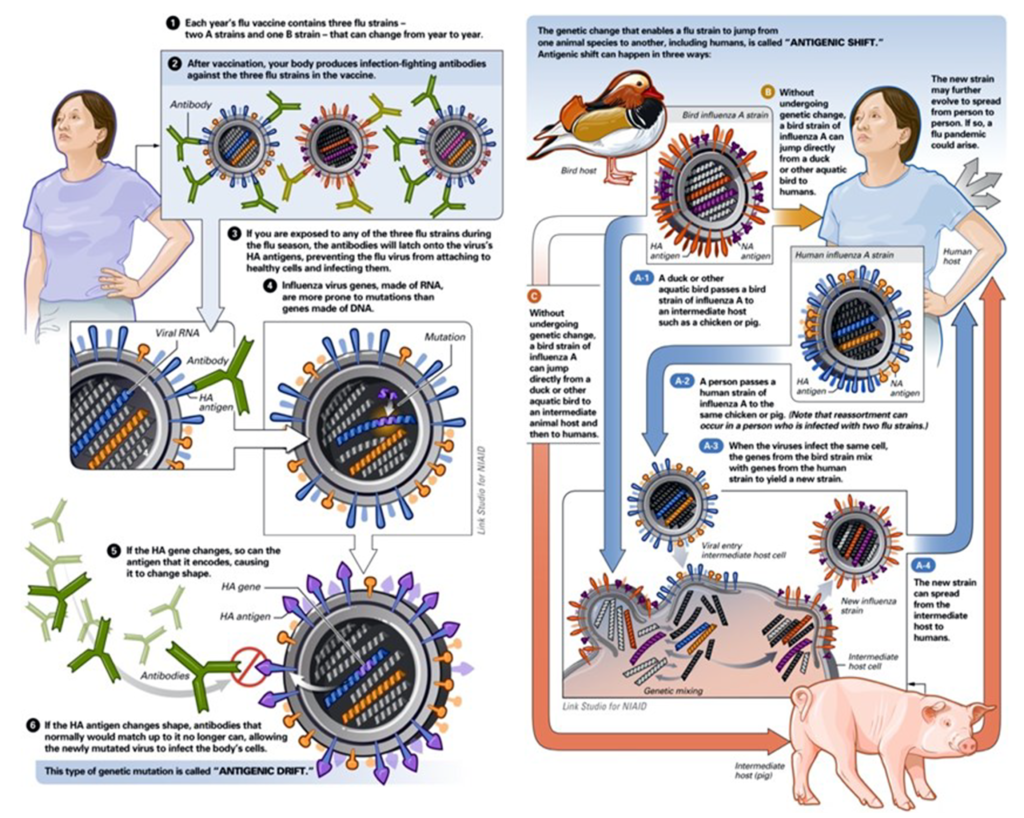
Influenza viruses are capable of evading the antibody-mediated immunity induced during previous infections or vaccinations by gradually accumulating mutations in HA and NA. This process, known as antigenic drift, necessitates frequent updates of influenza vaccines to ensure sufficient antigenic relatedness between the vaccine and emerging virus variants. Because HA is currently the main component of the inactivated influenza vaccines, human and animal influenza virus surveillance programmes are in place to monitor antigenic changes of the virus, primarily using the haemagglutination inhibition assay and viral RNA sequence analyses. Increased attention to antigenic drift of NA, measured using neuraminidase inhibition assays, could further increase the antigenic match between vaccines and circulating viruses. The antigenic evolution of influenza viruses was shown to be more rapid in human influenza viruses than in swine and equine viruses, which is presumably related to the size, mixing and age- structure (that is, humans have a longer lifespan than swine and horses) of the host populations.
Antigenic shift
In contrast to antigenic drift, antigenic shift refers to drastic changes in the antigenicity of the HA of circulating influenza A viruses; antigenic shift is associated with influenza A pandemics. The HA — and sometimes the NA — molecules of pandemic viruses are derived from antigenically diverse animal strains of influenza virus, which can be acquired by human influenza strains through reassortment. Viral reassortment is a more complex form of antigenic shift. It occurs when two viruses simultaneously infect the same animal. For example, pigs carry an endemic strain of influenza and can be infected with both human and avian influenza strains. The virus that caused the 2009 pandemic influenza (type A H1N1) is a quadruple reassortment virus. It contains genes from pigs normally found in Europe and Asia, avian–swine influenza genes, and human influenza genes.
Pandemic outbreaks are usually associated with the extinction of the previous circulating strains. However, in 1977, influenza A H1N1 viruses, not seen in humans since the 1957 influenza A H2N2 pandemic, started to co-circulate with influenza A H3N2 viruses. The 2009 influenza A H1N1 pandemic was caused by an influenza A H1N1 virus that was antigenically very different from the seasonal influenza A H1N1 virus circulating at the time and resulted in the extinction of the previous influenza A H1N1 human lineage, but it did not result in the extinction of the influenza A H3N2 viruses. Since 2009, influenza A H3N2 and influenza A H1N1 viruses derived from the 2009 pandemic virus and two lineages of influenza B virus are co-circulating in humans. Human influenza A virus infections with antigenically diverse avian H5N1, avian H7N9, swine H3N2 and other animal influenza viruses are constantly detected in geographical regions where these strains are prevalent owing to the contact of infected poultry or swine with humans.
However, no cases of sustained human-to-human transmission have been associated with these viruses, indicating that further adaptations need to take place for these viruses to become transmissible in humans.
Clinical Picture of avian influenza in Animals and Humans
In birds, fortunately, many avian influenza A viruses exhibit low pathogenicity, causing few signs of disease in infected wild birds. LPAI viruses are either asymptomatic or cause mild/subclinical disease (such as ruffled feathers and a drop in egg production) in chickens and poultry. However, these viruses are primed for continual emergence and pandemic potential, and some low-pathogenic viruses can mutate in poultry into highly pathogenic avian influenza viruses. HPAI viruses often cause severe disease and high mortality in infected poultry. HPAI A(H5) or A(H7) virus infections can cause disease that impacts multi-organ systems with mortality as high as 90-100% in chickens, often within 48 hours. HPAI A(H5N1) is one of the most contagious viruses occurring among birds, and the clinical outcome is typically deadly, especially in domestic poultry. However, ducks can be infected without any signs of illness. HPAI A(H5) and A(H7) virus infections in poultry can also spill back into wild birds, resulting in rapid geographic dissemination of the virus with bird migration. While some wild bird species can be infected with some HPAI A(H5) or A(H7) virus subtypes without appearing sick, other HPAI A(H5) and A(H7) virus subtypes can cause severe disease and mortality in some infected wild birds, as well as in infected poultry.

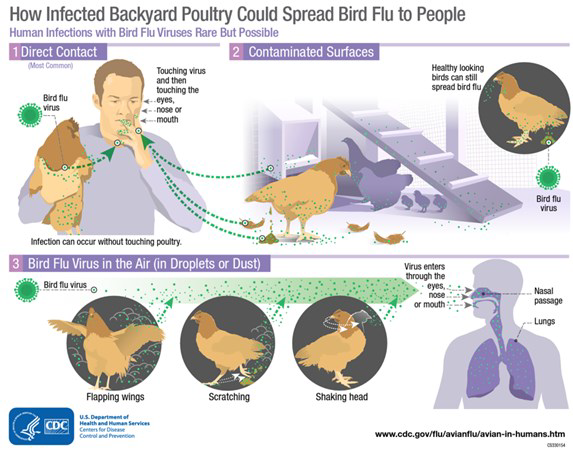
When birds are infected with this virus, they shed bird flu virus through their saliva, mucous and feces. Human infections are likely caused by accidental inoculation of virus into a person’s eyes, nose or mouth or via respiratory routes. Inhalation of virus occurs when the virus is in the air (in droplets or possibly dust) and a person breathes it in, or possibly when a person touches a fomite (contaminated object) that has virus on it then touches their mouth, eyes or nose. No human bird flu infections have been reported from proper handling of poultry meat or from eating properly cooked poultry or poultry products. Bird flu transmission from human–to–human is very rare, and when it has happened it has only spread to a few people. Human infections with bird flu viruses have ranged in severity from asymptomatic/mild illness to severe disease resulting in mortality. Despite there being a broad range of AI subtypes, fortunately, only a very select subset of these have been shown to infect humans with highly pathogenic consequences.
Asian lineage H7N9 and Asian lineage H5N1 viruses have been responsible for most human illness from bird flu virus-es globally, including the most serious illnesses and illness with the highest mortality. The emergence of the avian-derived H7N9 strain infecting humans was first described in March 2013 in China’s Yangtze River Delta. This viral sub-type is of a particular concern, as unlike H5N1, which is highly pathogenic in chickens and humans, H7N9 typically presents as an LPAI in chickens, but causes a high mortality rate in humans (40%), similar to that seen for H5N1 infections. H7N9 is one of several LPAI viruses in the H7 family capable of human infections, with viral transmission usually only acquired through close contact with host species. However, for reasons that are still unclear, H7N9 has greater transmissibility and more severe disease outcomes in humans than any other H7 viruses. The reported signs and symptoms of this type of infection in humans range from asymptomatic to mild (e.g., conjunctivitis/red eye) or mild, flu-like (e.g., upper respiratory symptoms), to more severe (e.g., pneumonia requiring hospitalization). Fever (temperature of 100oF [37.8oC] or greater), cough, sore throat, congestion, achiness, headaches, fatigue and shortness of breath or difficulty breathing are common. Diarrhea, nausea, vomiting or seizures may be observed more rarely.
Conclusion
The emergence of avian influenza viruses is of major concern to the avian and human population. The lack of pre-existing antibody immunity and their ability to cause severe disease through multiple host and viral mechanisms makes these viruses difficult to counter. Currently, these viruses are yet to effectively replicate and transmit between humans, however, experiments in ferrets show that only a few mutations are needed for H5N1 and H7N9 viruses to quickly adapt and become a major pandemic threat. Their ability to pass from birds to mammals commonly in contact with humans requires constant surveillance across all known bird reservoirs to limit the potential threat of an Avian influenza-derived pandemic. While H5N1 has been circulating among birds and poultry in various parts of the world for years, the latest outbreaks have sent warning signals to the scientific community, as the virus seems to be spreading to places previously unreached.
References:
Krammer F, Smith GJ, Fouchier RA, Peiris M, Kedzierska K, Doherty PC, Palese P, Shaw ML, Treanor J, Webster RG, García-Sastre A. Influenza (primer). Nature Reviews: Disease Primers. 2018;4(1):3.
Lycett SJ, Duchatel F, Digard P. A brief history of bird flu. Philosophical Transactions of the Royal Society B. 2019 Jun 24;374(1775):20180257.
https://www.cdc.gov/flu/avianflu/spotlights/2022-2023/h5n1-technical-report.htm
Dunning J, Thwaites RS, Openshaw PJ. Seasonal and pandemic influenza: 100 years of progress, still much to learn. Mucosal immunology. 2020 Jul 1;13(4):566-73.
Parums DV. Global Surveillance of Highly Pathogenic Avian Influenza Viruses in Poultry, Wild Birds, and Mammals to Prevent a Human Influenza Pandemic. Medical Science Monitor. 2023 Mar 1;29.
Sidik SM. How to stop the bird flu outbreak becoming a pandemic. Nature. 2023;615(7951):196-7.
https://asm.org/Articles/2022/July/Avian-Influenza-Past,-Present,-Future

Most Commented